
RESEARCH OVERVIEW
Our research program uses the tools of traditional organic chemistry together with Nature’s biological machinery to create small molecules and large interacting systems of molecules with novel chemical and biological properties. In the latter case we are creating living organisms with expanded genetic codes, evolving mitochondria-like organelles starting from bacterial endosymbionts inside yeast cells, and generating bacterial strains with chimeric RNA-DNA genomes. At the same time, we identify and characterize small molecules from phenotypic cell-based screens that affect a variety of normal and disease processes, including molecules that modulate the fate of endogenous somatic stem cells, selectively kill tumors and bacterial pathogens, and activate host protective mechanisms.
Synthetic Biology
The genetic codes of every known organism specify the same 20 amino acid building blocks using triplet codons generated from A, G, C and T. These twenty amino acids contain a limited number of functional groups including acids, amides, alcohols, basic amines and
thiols. Is this the ideal number or would additional amino acids allow the generation of proteins or even entire organisms with enhanced properties? The ability to site-specifically introduce amino acids with precisely tailored steric and electronic properties into proteins would also allow us to carry out "physical organic" studies of protein structure and function, much the same way as has been historically done with small molecules.
To this end, we have developed a methodology that allows one to genetically encode novel amino acids, beyond the common twenty, in both prokaryotic and eukaryotic organisms. This methodology involves the generation a unique codon-tRNA pair and corresponding aminoacyl-tRNA synthetase. Specifically, an orthogonal tRNA is constructed that is not a substrate for any natural aminoacyl synthetases and which inserts its cognate amino acid into the growing polypeptide chain in response to a nonsense or frameshift codon. A cognate synthetase is then generated which recognizes this unique tRNA and no other; the substrate specificity
of this synthetase is then evolved to recognize a desired "twenty first"amino acid, and no endogenous amino acid.
Recently we have also been attempting to recapitulate the endosymbiotic theory of mitochondrial evolution. It has been hypothesized that mitochondria evolved from a bacterial ancestor that initially became established in a host cell as an endosymbiont. We are testing this notion by engineering endosymbiosis between E. coli and S. cerevisiae. To this end it was necessary to engineer an E. coli strain
that expresses an ADP/ATP translocase that provides ATP to a respiration-deficient cox2 yeast mutant. In a reciprocal fashion, yeast provide thiamin to an endosymbiotic E. coli thiamin auxotroph. Expression of several SNARE-like proteins in E. coli was also required, likely to block lysosomal degradation of intracellular bacteria. Using this approach, we showed the presence of GFP-expressing E. coli endosymbionts inside the yeast cells by fluorescence microscopy and X-ray tomography. This readily manipulated system should allow experimental delineation of host-endosymbiont adaptations (for example, evolution of a reduced bacterial genome). We are also engineering other endosymbiotic systems, for example cyanobacterium-yeast chimeras.
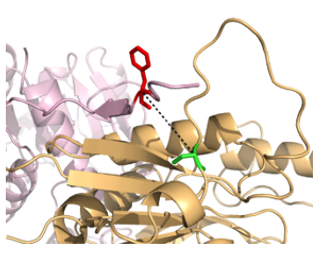
Figure 2: A noncanonical amino acid that stabilizes MetA from identified genetic selections
On another front we have begun to generate and characterize bacteria that contain unnatural nucleotides in their genome including modified bases and sugars. Almost five decades ago Crick, Orgel and others proposed that RNA might be able to support both genotype and phenotype. Since then, the RNA world hypothesis has been extensively investigated and the function of RNA templates has been studied in terms of evolution, replication and catalysis. We have used pathway engineering to generate strains of E. coli in which a
large fraction of 2’-deoxycytidine in the genome is substituted with the modified base 5-hydroxymethylcytidine. Surprisingly, mutant strains derived from these engineered bacteria show significant ribonucleotide content (approaching 50%) in their genomic templates. We have begun to characterize the chemical and biological properties of these chimeric templates and corresponding strains to determine the circumstances under which E. coli can incorporate ribonucleotides into its genome, and carry out replication, transcription, and repair with a chimeric genome. We are also pursuing alternative approaches to rationally engineer bacterial strains to incorporate ribonucleotides into their genomic templates. Such strains are likely to provide insights into the transition between the RNA and DNA worlds.engineered bacteria show significant ribonucleotide content (approaching 50%) in their genomic templates.
We have shown that this methodology can be used to efficiently incorporate a large number of noncanonical amino acids into proteins in E. coli, yeast and mammalian cells with fidelity and efficiency rivaling that of the common 20 amino acids. These include amino
Cell-Based Screens to Identify Small Molecules with Novel Biological Activities
We use "rationally designed" chemical libraries together with phenotypic and pathway-based screens to identify and characterize small molecules with novel biological activities. Chemistries have been developed to efficiently synthesize large combinatorial libraries of heterocyclic compounds designed around a large number of molecular scaffolds, including substituted purines, pyrimidines, quinazolines, pyrazines, pyrrolopyrimidine, pyrazolopyrimidine, phthalazines, pyridazines, and quinoxalines. These libraries are being screened in a large number of cell-based assays to identify molecules that both modulate and provide new insights into complex cellular processes, and which ultimately may lead to new therapeutic agents for the treatment of unmet human disease.
One area of interest is the identification of small molecules that affect stem cell fate. For example, a small molecule was found that expands hematopoietic stem cells (HSCs, the blood cell progenitor) and shown to act by antagonizing the aryl hydrocarbon receptor. On a related front a molecule was identified that selectively differentiates mesenchymal stem cells to chondrocytes by blocking the interaction of the chondrogenic transcription factor CBF-β with filamin A; a derivative of this molecule is has entered clinical trials for osteoarthritis. Similarly, we have found molecules that induce the formation of oligodendrocytes to remyelinate axons for the treatment of MS; induce β-cell formation for Type I diabetes; expand alveolar epithelial stem cells for lung disease and proliferate cardiomyocytes (by modulating Hippo-Yap signaling) for heart repair.
We have also used cell-based screens to identify molecules with anti-tumor and anti-bacterial activity and that protect against degenerative disease. For example, we have found molecules that block the mesenchymal-epithelial transition involved in cancer metastasis by binding to the cytoskeletal protein vimentin; and molecules that selectively kill glioblastoma tumor imitating cells by binding and modulating the ubiquitination of RIPK2. On another front we have found molecules that protect against oxidative stress involved in aging and neurodegenerative diseases by activating the anti-oxidant response pathway through a novel posttranslational modification. In addition, we have ongoing programs aimed at a number of neglected disease including tuberculosis, malaria, viral and helminth infections. Many of these programs are being carried out in collaboration with scientists at Calibr which has state of the art HT screening facilities, compound libraries, medicinal and protein chemistry and pharmacology.
Importantly, we investigate the mechanisms of these molecules in order to gain new insights into the complex biology of normal and diseased cell physiology. We use a combination of biochemical and genomic techniques including cDNA complementation, siRNA knockdown, photoaffinity labeling and peptide mapping, phosphoproteome and mRNA expression analysis, as well as medicinal chemistry and pharmacology to assess the activity of these molecules in relevant in vitro and in vivo disease models. Using these methods, we have shown that these molecules can block or activate key developmental pathways, selectively modulate nuclear localization of transcriptional activators, antagonize nuclear receptors and GPCRs, block protein degradation or interfere with protein-protein interactions.
acids with novel steric/packing and electronic properties; photocrosslinking amino acids; bio-orthogonal keto, acetylene, azide, and boronate containing amino acids which can be used to selectively modify proteins with biophysical probes, drugs, and novel functional groups; photocaged and photoisomerizable amino acids; metal binding amino acids; and amino acids that contain fluorescent or IR active side chains to probe protein structure and dynamics. We have used this technology to synthesize structurally defined bispecific antibodies, antibody-drug conjugates and vaccines as novel therapeutics; to generate novel biological probes (photocrosslinking agents, spectroscopic probes, and photocaged proteins) to explore protein structure and function in vitro and in vivo; and to rationally design or evolve peptides and proteins with enhanced catalytic activity, thermal stability or binding properties. We have also shown that one can introduce noncanonical amino acidsinto hematopoietic stem cells and their progeny in mice, as well as "synthesize" a completely autonomous bacterium that not only genetically encodes a novel amino acid, but also biosynthesizes this amino acid from basic carbon sources.

Figure 4: Metabolic engineering to produce E. coli with altered genomes
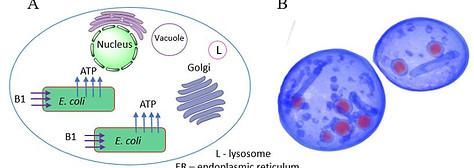
Figure 3: Recapitulating mitochondrial evolution starting from bacterial endosymbionts in yeast cells.
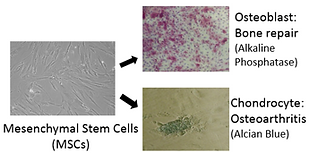
Figure 5: Small molecules that control the fate of endogenous multipotent stem cells
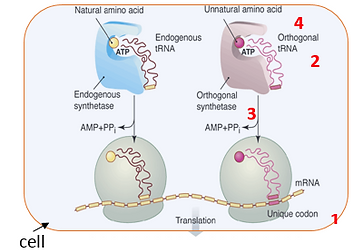
Figure 1: Structure-based design and in vitro evolution tools are used to reprogram the ribosomal machinery to expand genetic code
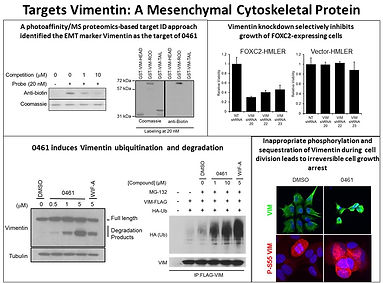
Figure 6: A small molecule block the epithelial to mesenchymal transition in tumor metastasis by binding vimentin
